Robust Brain Tissue Segmentation in AD Using Comparative Linear Transformation and Deep Learning
Main Article Content
Abstract
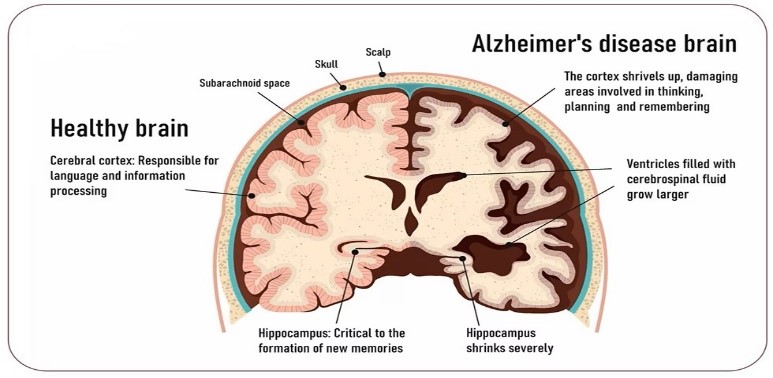
As a progressive neurological disease, Alzheimer's disease (AD), if no preventative measures are taken, can result in dementia and a severe decline in brain function, making it difficult to perform basic tasks. Over 1 in 9 people suffer from dementia caused by Alzheimer's disease and require uncompensated care. The hippocampus is extracted from MRI scans of the brain via image segmentation have been useful for diagnosing Alzheimer's disease (AD).The segmentation of the CSF region in brain MRI is critical for analyzing the stages of AD. The extraction of Hippocampus from an MRI of the brain is greatly influenced by the contrast of the images. Using comparative linear transformation in the horizontal and vertical dimensions as well as statistical edge-based features, this article proposes a robust method for segmentation technique for the extraction of Hippocampus from brain MRI. These transformations aid in balancing the brain image's thin and dense fluid extractions. Through use of the ADNI dataset, the proposed approach had a 99% success rate in segmentation.
Article Details
References
Sperling RA, Aisen PS, Beckett LA, et al. “Toward defining the preclinical stages of Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease”. Alzheimers Dement.2011;7:280–92.
Albert MS, DeKosky ST, Dickson D, et al. “The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease”. Alzheimers Dement. 2011;7:270–9.
McKhann GM, Knopman DS, Chertkow H, et al. “The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging and the Alzheimer’s Association workgroup”. Alzheimers Dement. 2011;7(3):263–9.
Peskind ER, Riekse R, Quinn JF, et al. “Safety and acceptability of the research lumbar puncture”. Alzheimer Dis Assoc Disord. 2005;19(4):220–5.
Lewczuk P, Riederer P, O'Bryant SE, et al. “Cerebrospinal fluid and blood biomarkers for neurodegenerative dementias: an update of the Consensus of the Task Force on Biological Markers in Psychiatry of the World Federation of Societies of Biological Psychiatry.” World J Biol Psychiatry. 2017: 1–85. https://doi.org/10.1080/15622975.2017.1375556 [Epub ahead of print].
Fagan AM, Perrin RJ. “Upcoming candidate cerebrospinal fluid biomarkers of Alzheimer’s disease”. Biomark Med. 2012;6(4):455–76.
Lewczuk P, Kornhuber J. “Neurochemical dementia diagnostics in Alzheimer’s disease: where are we now and where are we going?” Expert Rev Proteomics. 2011;8(4):447–58.
Hampel H, Burger K, Teipel SJ, et al.” Core candidate neurochemical and imaging biomarkers of Alzheimer’s disease”. Alzheimers Dement. 2008;4:38–48.
Hansson O, Zetterberg H, Buchhave P, et al. “Association between CSF biomarkers and incipient Alzheimer’s disease in patients with mild cognitive impairment: a follow-up study”. Lancet Neurol. 2006;5:228–34.
Mattsson N, Zetterberg H, Hansson O, et al. ‘CSF biomarkers and incipient Alzheimer disease in patients with mild cognitive impairment’. JAMA. 2009; 302:385–93.
Petersen RC.” Mild cognitive impairment as a diagnostic entity”. J Intern Med. 2004;256(3):183–94.
O. Merveille, B. Naegel, H. Talbot, N. Passat, ???? d “variational restoration of curvilinear structures with prior-based directional regularization”, IEEE Transactions on Image Processing 28 (8) (2019) 3848– 3859.
Gao F et al (2022) “A combination model of AD biomarkers revealed by machine learning precisely predicts Alzheimer’s dementia: China Aging and Neurodegenerative Initiative (CANDI) study”. Alzheimers Dement. https://doi.org/10.1002/alz. 12700
S. Kumar, A. Adarsh, B. Kumar, A. K. Singh, “An automated early diabetic retinopathy detection through improved blood vessel and optic disc segmentation”, Optics & Laser Technology 121 (2020) 105815
A. Mosinska, M. Kozi?ski, P. Fua, “Joint segmentation and path classification of curvilinear structures”, IEEE transactions on pattern analysis and machine intelligence 42 (6) (2019) 1515–1521.
J. Xie, L. Fang, B. Zhang, J. Chanussot, S. Li, “Super resolution guided deep network for land cover classification from remote sensing images”, IEEE Transactions on Geoscience and Remote Sensing 60 (2021) 1–12
K. Blennow, H. Hampel, M. Weiner, H. Zetterberg, “Cerebrospinal fluid and plasma biomarkers in Alzheimer disease”, Nat. Rev. Neurol. 6 (2010) 131–144, https://doi. org/10.1038/nrneurol.2010.
P. Lewczuk, P. Riederer, S.E. O'Bryant, M.M. Verbeek, B. Dubois, P.J. Visser, K.A. Jellinger, S. Engelborghs, A. Ramirez, L. Parnetti, C.R. Jack, C.E. Teunissen, H. Hampel, A. Lleó, F. Jessen, L. Glodzik, M.J. de Leon, A.M. Fagan, J.L. Molinuevo, W.J. Jansen, B. Winblad, L.M. Shaw, U. Andreasson, M. Otto, B. Mollenhauer, J. Wiltfang, M.R. Turner, I. Zerr, R. Handels, A.G. Thompson, G. Johansson, N. Ermann, J.Q. Trojanowski, I. Karaca, H. Wagner, P. Oeckl, L. van Waalwijk VanDoorn, M. Bjerke, D. Kapogiannis, H.B. Kuiperij, L. Farotti, Y. Li, B.A. Gordon, S. Epelbaum, S.J.B. Vos, C.J.M. Klijn, W.E. Van Nostrand, C. Minguillon, M. Schmitz, C. Gallo, A. Lopez Mato, F. Thibaut, S. Lista, D. Alcolea, H. Zetterberg, K. Blennow, J. Kornhuber, “Cerebrospinal fluid and blood biomarkers for neurode- generative dementias: an update of the consensus of the task force on biological markers in psychiatry of the world federation of societies of biological psychiatry”, World J. Biol. Psychiatry 19 (2018) 244–328, https://doi.org/10.1080/15622975. 2017.1375556.
S. Janelidze, H. Zetterberg, N. Mattsson, S. Palmqvist, H. Vanderstichele, O. Lindberg, D. Van Westen, E. Stomrud, L. Minthon, K. Blennow, “CSF Ab42/Ab40 and Ab42/Ab38 ratios: better diagnostic markers of Alzheimer disease”, Ann. Clin. Transl. Neurol. (3) (2016) 154–165, https://doi.org/10.1002/acn3.274
Ana Oliveira, Yosef Ben-David, Susan Smit , Elena Popova, Milica Mili?. Machine Learning for Decision Optimization in Complex Systems. Kuwait Journal of Machine Learning, 2(3). Retrieved from http://kuwaitjournals.com/index.php/kjml/article/view/201
J. Toombs, R.W. Paterson, M.P. Lunn, J.M. Nicholas, N.C. Fox, M.D. Chapman, J.M. Schott, H. Zetterberg, “Identification of an important potential confound in CSF AD studies: Aliquot volume”, Clin. Chem. Lab. Med. 51 (2013) 2311–2317, https:// doi.org/10.1515/cclm-2013-0293.
C. Gervaise-Henry, G. Watfa, E. Albuisson, A. Kolodziej, B. Dousset, J.L. Olivier, T.R. Jonveaux, C. Malaplate-Armand, “Cerebrospinal fluid A?42/A?40 as a means to limiting tube- and storage-dependent pre-analytical variability in clinical setting”, J. Alzheimers Dis. 57 (2017) 437–445, https://doi.org/10.3233/JAD-160865.
E. Willemse, K. van Uffelen, B. Brix, S. Engelborghs, H. Vanderstichele, C. Teunissen, “How to handle adsorption of cerebrospinal fluid amyloid ? (1–42) in laboratory practice? Identifying problematic handlings and resolving the issue by use of the A?42/A?40ratio”, Alzheimers Dement. 13 (2017) 885–892, https://doi.org/10.1016/j.jalz.2017.01.010.
Srhoj-Egekher, V.; Benders, M.; Viergever, M.A.; Išgum, I. “Automatic neonatal brain tissue segmentation with MRI”. SPIE Med. Imaging 2013, 8669, 86691K. [CrossRef]
Anbeek, P.; Išgum, I.; Van Kooij, B.J.M.; Mol, C.P.; Kersbergen, K.J.; Groenendaal, F.; Viergever, M.A.; De Vries, L.S.; Benders, M. “Automatic Segmentation of Eight Tissue Classes in Neonatal Brain MRI.” PLoS ONE 2013, 8, e81895. [CrossRef]
Ashraf Dhannon Hasan, Abbas Salim Kadhim, Mustafa A. Ali. (2023). Medical Image Compression Using Hybrid Compression Techniques. International Journal of Intelligent Systems and Applications in Engineering, 11(4s), 634–648. Retrieved from https://ijisae.org/index.php/IJISAE/article/view/2741
Vrooman, H.; Cocosco, C.; Lijn, F.; Stokking, R.; Ikram, M.; Vernooij, M.; Breteler, M.; Niessen, W.J. “Multi-spectral brain tissue segmentation using automatically trained k-nearestneighbor classification”. NeuroImage 2007, 37, 71–81. [CrossRef] [PubMed]
Makropoulos, A.; Gousias, I.S.; Ledig, C.; Aljabar, P.; Serag, A.; Hajnal, J.V.; Edwards, A.D.; Counsell, S.; Rueckert, D. “Automatic Whole Brain MRI Segmentation of the Developing Neonatal Brain”, IEEE Trans. Med. Imaging 2014, 33, 1818–1831. [CrossRef] [PubMed]
Christensen, G.E.; Rabbitt, R.D.; Miller, M.I. “Deformable templates using large deformation kinematics.” IEEE Trans. Image Process. 1996, 5, 1435–1447. [CrossRef]
Davatzikos, C.; Prince, J. “Brain image registration based on curve mapping”. In Proceedings of the IEEE Workshop on Biomedical Image Analysis, Seattle, WA, USA, 24–25 June 1994; pp. 245–254.
Carmichael, O.T.; Aizenstein, H.J.; Davis, S.W.; Becker, J.T.; Thompson, P.M.; Meltzer, C.C.; Liu, Y. “Atlas-based hippocampus segmentation in Alzheimer’s disease and mild cognitive impairment”. NeuroImage 2005, 27, 979–990. [CrossRef]
Wang, L.; Gao, Y.; Shi, F.; Li, G.; Gilmore, J.; Lin, W.; Shen, D. Links: “Learning-based multi-source integration framework for segmentation of infant brain images”. NeuroImage 2014, 108, 734–746.
Chi¸ta, S.M.; Benders, M.; Moeskops, P.; Kersbergen, K.J.; Viergever, M.A.; Išgum, I. “Automatic segmentation ? of the preterm neonatal brain with MRI using supervised classification”. SPIE Med Imaging 2013, 8669, 86693.
Cuingnet, R.; Gerardin, E.; Tessieras, J.; Auzias, G.; Lehéricy,S.; Habert, M.-O.; Chupin, M.; Benali, H.; Colliot, O. “Automatic classification of patients with Alzheimer’s disease from structural MRI: A comparison of ten methods using the ADNI database”. NeuroImage 2011, 56, 766–781. [CrossRef]
Wolz, R.; Julkunen, V.; Koikkalainen, J.; Niskanen, E.; Zhang, D.P.; Rueckert, D.; Soininen, H.; Lötjönen, J.M.P.; “The Alzheimer’s Disease Neuroimaging Initiative. Multi-Method Analysis of MRI Images in Early Diagnostics of Alzheimer’s Disease”. PLoS ONE 2011, 6, e25446. [CrossRef] [PubMed]
Braak, H.; Braak, E. “Staging of alzheimer’s disease-related neurofibrillary changes”. Neurobiol. Aging 1995, 16, 271–278. [CrossRef]
Dubois, B.; Feldman, H.H.; Jacova, C.; DeKosky, S.T.; Barberger-Gateau, P.; Cummings, J.; Delacourte, A.; Galasko, U.; Gauthier, S.; Jicha, G.; et al. Research criteria for the diagnosis of Alzheimer’s disease: Revising the NINCDS–ADRDA criteria. Lancet Neurol. 2007, 6, 734–746. [CrossRef]
T. Shi, N. Boutry, Y. Xu, T. Géraud,” Local intensity order transformation for robust curvilinear object segmentation”, IEEE Transactions on Image Processing 31 (2022) 2557–2569
D. Nie et al., “Fully convolutional networks for multi-modality isointense infant brain image segmentation”, Procedding of the 2016 IEEE International Symposium on Biomedical Imaging (ISBI), pp. 1342-1345, 2016.
T. Shi, N. Boutry, Y. Xu, T. Géraud, “Local intensity order transformation for robust curvilinear object segmentation”, IEEE Transactions on Image Processing 31 (2022) 2557–2569.
K. Acharya, D. Ghoshal, “Central moment and multinomial based sub image clipped histogram equalization for image enhancement”, International Journal of Image, Graphics and Signal Processing (IJIGSP) 13 (1) (2021) 1–12
M. Zhu, K. Zeng, G. Lin, Y. Gong, T. Hao, K. Wattanachote, X. Luo, “Iternet++: An improved model for retinal image segmentation by curvelet enhancing, guided filtering, offline hard-sample mining, and test-time augmenting”, IET Image Processing (2022).