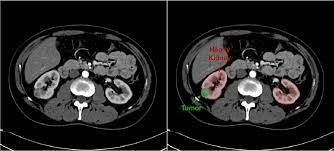
U-Capkidnets++-: A Novel Hybrid Capsule Networks with Optimized Deep Feed Forward Networks for an Effective Classification of Kidney Tumours Using CT Kidney Images
Main Article Content
Abstract
Chronic Kidney Diseases (CKD) has become one among the world wide health crisis and needs the associated efforts to prevent the complete organ damage. A considerable research effort has been put forward onto the effective seperation and classification of kidney tumors from the kidney CT Images. Emerging machine learning along with deep learning algorithms have waved the novel paths of tumor detections. But these methods are proved to be laborious and its success rate is purely depends on the previous experiences. To achieve the better classification and segmentation of tumors, this paper proposes the hybrid ensemble of visual capsule networks in U-NET deep learning architecture and w deep feed-forward extreme learning machines. The proposed framework incorporates the data-preprocessing powerful data augmentation, saliency tumor segmentation (STS) followed by the classification phase. Furthermore, classification levels are constructed based upon the feed forward extreme learning machines (FFELM) to enhance the effectiveness of the suggested model .The extensive experimentation has been conducted to evaluate the efficacy of the recommended structure and matched with the other prevailing hybrid deep learning model. Experimentation demonstrates that the suggested model has showed the superior predominance over the other models and exhibited DICE co-efficient of kidney tumors as high as 0.96 and accuracy of 97.5 %respectively.
Article Details
References
Abraham N, Khan NM. A novel focal tversky loss function with improved attention u-net for lesion segmentation. In: 2019 IEEE 16th international symposium on biomedical imaging (ISBI 2019). IEEE; 2019. p. 683–7.
Badrinarayanan V, Kendall A, Cipolla R. Segnet: a deep convolutional encoderdecoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell 2017;39(12):2481–95
Cairns P. Renal cell carcinoma. Canc Biomarkers 2011;9(1–6):461–73
Christ PF, Ettlinger F, Grün F, Elshaera MEA, Lipkova J, Schlecht S, Ahmaddy F, Tatavarty S, Bickel M, Bilic P, et al. Automatic liver and tumor segmentation of ct and mri volumes using cascaded fully convolutional neural networks. 2017. arXiv preprint arXiv:1702.05970.
Çiçek O, € Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3d u-net: learning dense volumetric segmentation from sparse annotation. In: International conference on medical image computing and computer-assisted intervention. Springer; 2016. p. 424–32
Dai J, Qi H, Xiong Y, Li Y, Zhang G, Hu H, Wei Y. Deformable convolutional networks. In: Proceedings of the IEEE international conference on computer vision; 2017. p. 764–73
Fasihi MS, Mikhael WB. Overview of current biomedical image segmentation methods. In: 2016 international conference on computational science and computational intelligence (CSCI). IEEE; 2016. p. 803–8.
Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016;278(2):563–77.
Grana C, Borghesani D, Cucchiara R. Optimized block-based connected components labeling with decision trees. IEEE Trans Image Process 2010;19(6): 1596–609
Hatamizadeh A, Terzopoulos D, Myronenko A. Boundary aware networks for medical image segmentation. 2019. arXiv preprint arXiv:1908.08071
G. Chen et al., "Prediction of Chronic Kidney Disease Using Adaptive Hybridized Deep Convolutional Neural Network on the Internet of Medical Things Platform," in IEEE Access, vol. 8, pp. 100497-100508, 2020, doi: 10.1109/ACCESS.2020.2995310.
N. Bhaskar and S. Manikandan, "A Deep-Learning-Based System for Automated Sensing of Chronic Kidney Disease," in IEEE Sensors Letters, vol. 3, no. 10, pp. 1-4, Oct. 2019, Art no. 7001904, doi: 10.1109/LSENS.2019.2942145.
J. Qin, L. Chen, Y. Liu, C. Liu, C. Feng and B. Chen, "A Machine Learning Methodology for Diagnosing Chronic Kidney Disease," in IEEE Access, vol. 8, pp. 20991-21002, 2020, doi: 10.1109/ACCESS.2019.2963053.
L. Antony et al., "A Comprehensive Unsupervised Framework for Chronic Kidney Disease Prediction," in IEEE Access, vol. 9, pp. 126481-126501, 2021, doi: 10.1109/ACCESS.2021.3109168.
D. Pavithra and R. Vanithamani, "Chronic Kidney Disease Detection from Clinical Data using CNN," 2021 IEEE International Conference on Distributed Computing, VLSI, Electrical Circuits and Robotics (DISCOVER), 2021, pp. 282-285, doi: 10.1109/DISCOVER52564.2021.9663670.
G. S. K. G. Prasad, A. A. Chowdari, K. P. Jona and R. Senapati, "Detection of CKD from CT Scan images using KNN algorithm and using Edge Detection," 2022 2nd International Conference on Emerging Frontiers in Electrical and Electronic Technologies (ICEFEET), 2022, pp. 1-4, doi: 10.1109/ICEFEET51821.2022.9848173.
N. Bhaskar, M. Suchetha and N. Y. Philip, "Time Series Classification-Based Correlational Neural Network With Bidirectional LSTM for Automated Detection of Kidney Disease," in IEEE Sensors Journal, vol. 21, no. 4, pp. 4811-4818, 15 Feb.15, 2021, doi: 10.1109/JSEN.2020.3028738.
S. Akter et al., "Comprehensive Performance Assessment of Deep Learning Models in Early Prediction and Risk Identification of Chronic Kidney Disease," in IEEE Access, vol. 9, pp. 165184-165206, 2021, doi: 10.1109/ACCESS.2021.3129491.
D. Weerasinghe, B. T. G. S. Kumara, B. Kuhaneswaran and S. K. Gunathilake, "Identifying the Type of Chronic Kidney Disease Based on Heavy Metals in Soil using ANN," 2021 Third International Sustainability and Resilience Conference: Climate Change, 2021, pp. 270-274, doi: 10.1109/IEEECONF53624.2021.9667974.
V. Shanmugarajeshwari and M. Ilayaraja, "Chronic Kidney Disease for Collaborative Healthcare Data Analytics using Random Forest Classification Algorithms," 2021 International Conference on Computer Communication and Informatics (ICCCI), 2021, pp. 1-14, doi: 10.1109/ICCCI50826.2021.9402574.
Soo-Chang Pei; Yu-Zhe Hsiao,” Spatial Affine transformations of images by using fractional shift fourier transform”, 2015 IEEE International Symposium on Circuits and Systems (ISCAS), doihttps://doi.org/10.1109/ISCAS.2015.7168951
Banerjee S, Mitra S, Shankar BU, Hayashi Y (2016) A novel GBM saliency detection model using multi-channel MRI. Plos one 11(1):e0146388
Takacs P, Manno-Kovacs A (2018) Mri brain tumor segmentation combining saliency and convolutional ´ network features. In: 2018 International conference on content-based multimedia indexing (CBMI), pp 1–6
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 234–241
G.-B. Huang, Q.-Y. Zhu, and C.-K. Siew, “Extreme learning machine: theory and applications,” Neurocomputing, vol. 70, no. 1, pp. 489–501, 2006.
Wang B, Huang S, Qiu J, et al. Parallel online sequential extreme learning machine based on MapReduce. Neurocomputing 2015; 149: 224-32.