A Probabilistic Adaptive Cerebral Cortex Segmentation Algorithm for Magnetic Resonance Human Head Scan Images
Main Article Content
Abstract
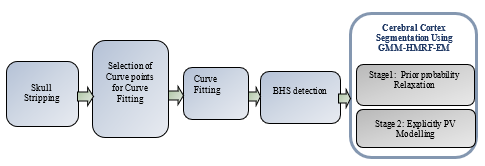
The total efficiency of Magnetic Resonance Imaging (MRI) results in the need for human involvement in order to appropriately detect information contained in the image. Currently, there has been a surge in interest in automated algorithms that can more precisely divide medical image structures into substructures than prior attempts. Instant segregation of cerebral cortex width from MRI scanned images is difficult due to noise, Intensity Non-Uniformity (INU), Partial Volume Effects (PVE), MRI's low resolution, and the very complicated architecture of the cortical folds. In this paper, a Probabilistic Adaptive Cerebral Cortex Segmentation (PACCS) approach is proposed for segmenting brain areas of T1 weighted MRI of human head images. Skull Stripping (SS), Brain Hemisphere Segmentation (BHS) and CCS are the three primary processes in the suggested technique. In step 1, Non-Brain Cells (NBC) is eliminated by a Contour-Based Two-Stage Brain Extraction Method (CTS-BEM). Step 2 details a basic BHS technique for Curve Fitting (CF) detection in MRI human head images. The left and right hemispheres are divided using the discovered Mid-Sagittal Plane (MSP). At last, to enhance a probabilistic CCS structure with adjustments such as prior facts change to remove segmentation bias; the creation of express direct extent training; and a segmentation version based on a regionally various Gaussian Mixture Model- Hidden Markov Random Field – Expectation Maximization (GMM-HMRF-EM). The underlying partial extent categorization and its interplay with found image intensities are represented as a spatially correlated HMRF within the GMM-HMRF-EM method. The proposed GMM-HMRF method estimates HMRF parameters using the EM technique. Finally, the outcomes of segmentation are evaluated in terms of precision, recall, specificity, Jaccard Similarity (JS), and Dice Similarity (DS). The proposed method works better and more consistently than the present locally Varying MRF (LV-MRF), according to the experimental findings obtained by using the suggested GMM-HMRF-EM methodology to 18 individuals' brain images.
Article Details
References
M.A. Balafar, “Gaussian mixture model based segmentation methods for brain MRI images,” Artificial Intelligence Review, vol.41, no.3, pp.429-439, 2014, doi:10.1007/s10462-012-9317-3.
A. A. Pravitasari, Y. P. Hermanto, N. Iriawan, I. Irhamah, K. Fithriasari, S. W. Purnami and W. Ferriastuti, “MRI-based brain tumor segmentation using Gaussian mixture model with reversible jump Markov chain Monte Carlo algorithm,” In AIP Conference Proceedings, vol. 2194, no. 1, AIP Publishing, 2019, December, doi:10.1063/1.5139817 .
W. Zheng, J.P. Kim, M. Kadbi, B. Movsas, I.J. Chetty and C.K. Glide-Hurst, “Magnetic resonance–based automatic air segmentation for generation of synthetic computed tomography scans in the head region,” International Journal of Radiation Oncology* Biology* Physics, vol.93, no.3, pp.497-506, 2015, doi:10.1016/j.ijrobp.2015.07.001.
K. Keraudren, M. Kuklisova-Murgasova, V. Kyriakopoulou, C. Malamateniou, M. A. Rutherford, B. Kainz and D. Rueckert, “Automated fetal brain segmentation from 2D MRI slices for motion correction,” NeuroImage, vol. 101, pp. 633-643, 2014, doi:10.1016/j.neuroimage.2014.07.023.
A. Jog, A. Hoopes, D. N. Greve, K. Van Leemput and B. Fischl, “PSACNN: Pulse sequence adaptive fast whole brain segmentation,” NeuroImage, vol. 199, pp. 553-569, 2019, doi:10.1016/j.neuroimage.2019.05.033.
Q. Mahmood, A. Chodorowski, and M. Persson, “Automated MRI brain tissue segmentation based on mean shift and fuzzy c-means using a priori tissue probability maps,” Irbm, vol. 36, no. 3, pp. 185-196, 2015, doi:10.1016/j.irbm.2015.01.007.
B. Lee, N. Yamanakkanavar, and J. Y. Choi, “Automatic segmentation of brain MRI using a novel patch-wise U-net deep architecture,” Plos one, vol. 15, no. 8, pp. e0236493, 2020, doi:10.1371/journal.pone.0236493.
T.A. Tuan, J.Y. Kim and P.T. Bao, “3D brain magnetic resonance imaging segmentation by using bitplane and adaptive fast marching,” International Journal of Imaging Systems and Technology, vol.28, no.3, pp.223-230, 2018, doi:10.1002/ima.22273.
K.L. Perdue and S.G. Diamond, “T1 magnetic resonance imaging head segmentation for diffuse optical tomography and electroencephalography”, Journal of biomedical optics, vol.19, no.2, pp.1-8, 2014, doi:10.1117/1.JBO.19.2.026011.
J. Lai, H. Zhu and X. Ling, “Segmentation of brain mr images by using fully convolutional network and gaussian mixture model with spatial constraints,” Mathematical Problems in Engineering, 2019, doi:10.1155/2019/4625371.
Y. Song, Z. Ji, Q. Sun, “An extension Gaussian mixture model for brain MRI segmentation,” In 2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (pp. 4711-4714). IEEE 2014, August, doi:10.1109/EMBC.2014.6944676.
Kartika S. (2016). Analysis of “SystemC” design flow for FPGA implementation. International Journal of New Practices in Management and Engineering, 5(01), 01 - 07. Retrieved from http://ijnpme.org/index.php/IJNPME/article/view/41.
J. Qiao, X. Cai, Q. Xiao, Z. Chen, P. Kulkarni, C. Ferris and S. Sridhar, “Data on MRI brain lesion segmentation using K-means and Gaussian Mixture Model-Expectation Maximization,” Data in brief, vol. 27, 104628, 2019, doi:10.1016/j.dib.2019.104628.
P. Moeskops, M. A. Viergever, A. M. Mendrik, L. S. De Vries, M. J. Benders and I. Išgum, “Automatic segmentation of MR brain images with a convolutional neural network,” IEEE transactions on medical imaging, vol. 35, no. 5, pp. 1252-1261, 2016, doi:10.1109/TMI.2016.2548501.
T. A. Tuan, J. Y. Kim and P. T. Bao, “Adaptive region growing for skull, brain, and scalp segmentation from 3d mri,” Biomedical Engineering: Applications, Basis and Communications, vol. 31, no. 05, pp. 1950033, 2019, doi:10.4015/S1016237219500339.
M. J. Cardoso, M. J. Clarkson, G. R. Ridgway, M. Modat, N. C. Fox, S. Ourselin and Alzheimer's Disease Neuroimaging Initiative. “ LoAd: a locally adaptive cortical segmentation algorithm,” NeuroImage, 56(3), 1386-1397, 2011, doi:10.1016/j.neuroimage.2011.02.013.
Kim, H. ., Jeong, Y. ., Seo, S. ., Youn, J. ., & Lee, D. (2023). A Study on Radiant Heat Application to the Curing Process for Improvement of Free-Form Concrete Panel Productivity. International Journal of Intelligent Systems and Applications in Engineering, 11(4s), 157–164. Retrieved from https://ijisae.org/index.php/IJISAE/article/view/2583.
K. Somasundaram and P. Kalavathi, “Medical image contrast enhancement based on gamma correction. Int J Knowl Manag e-learning, vol. 3, no. 1, pp. 15-18, 2011.
K. Somasundaram and P. Kalavathi, “Medical image binarization using square wave representation,” In International Conference on Logic, Information, Control and Computation, pp. 152-158, Berlin, Heidelberg: Springer Berlin Heidelberg, 2011, February, doi:10.1007/978-3-642-19263-0_19.
Q. Wang, “GMM-based hidden Markov random field for color image and 3D volume segmentation,” arXiv preprint arXiv:1212.4527, 2012, doi:10.48550/arXiv.1212.4527.
P. Kalavathi and V. B. Surya Prasath, “Automatic segmentation of cerebral hemispheres in MR human head scans,” International Journal of Imaging Systems and Technology, vol. 26, no. 1, pp. 15-23, 2016, doi:10.1002/ima.22152.
K. Somasundaram and P. Kalavathi, “Contour-based brain segmentation method for magnetic resonance imaging human head scans,” Journal of computer assisted tomography, vol. 37, no. 3, pp. 353-368, 2013, doi: 10.1097/RCT.0b013e3182888256.
Luca Ferrari, Deep Learning Techniques for Natural Language Translation , Machine Learning Applications Conference Proceedings, Vol 2 2022.
T. Rohlfing, “Image similarity and tissue overlaps as surrogates for image registration accuracy: widely used but unreliable,” IEEE transactions on medical imaging, vol. 31, no. 2, pp. 153-163, 2011, doi:10.1109/TMI.2011.2163944.
D. Izquierdo-Garcia, A. E. Hansen, S. Förster, D. Benoit, S. Schachoff, S. Fürst and C. Catana, “An SPM8-based approach for attenuation correction combining segmentation and nonrigid template formation: application to simultaneous PET/MR brain imaging,” Journal of Nuclear Medicine, vol. 55, no. 11, pp. 1825-1830, 2014, doi:10.2967/jnumed.113.136341.
L. Palumbo, P. Bosco, M. E. Fantacci, E. Ferrari, P. Oliva, G. Spera and A. Retico, “Evaluation of the intra-and inter-method agreement of brain MRI segmentation software packages: A comparison between SPM12 and FreeSurfer v6. 0.,” Physica Medica, vol. 64, pp. 261-272, 2019, doi:10.1016/j.ejmp.2019.07.016.