A Novel Densenet-324 Densely Connected Convolution Neural Network for Medical Crop Classification using Remote Sensing Hyperspectral Satellite Images
Main Article Content
Abstract
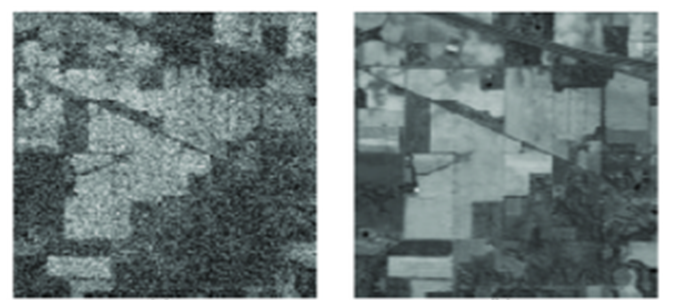
In the past few decades, importance of the medicinal Crops is extending to a large extent due to its benefits in treating life-threatening diseases. Medicinal Crop has excellent medicinal properties on its roots, stem, and leaves to prevent human and animal health. Particularly detection and identification of the Crop classes are effectively carried out using hyperspectral images as discrimination of the target feature or objects is simple and it contains rich information containing the spatial and temporal details of underlying the land cover. However, Crop classification using machine learning architectures concerning spectral characteristics obtained on the anatomical features and morphological features. Extracted features towards classification lead to several challenges such as large spatial and temporal variability and spectral signatures similarity between different objects. A further hyperspectral image poses several difficulties with changes in illumination, environment, and atmospheric aspects. To tackle those non-trivial challenges, DenseNet-324 Densely Connected convolution neural network architecture has been designed in this work to discriminate the crop and medical Crop effectively in the interested areas. Initially, the Hyperspectral image is pre-processed against a large number of noises through the employment of the noise removal technique and bad line replacement techniques. Pre-processed image is explored to image segmentation using the global thresholding method to segment it into various regions based on spatial pieces of information on grouping the neighboring similar pixels intensity or textures. Further regions of the image are processed using principle component analysis to extract spectral features of the image. That extracted feature is employed to ant colony optimization technique to obtain the optimal features. Computed optimal features are classified using Convolution Neural Network with a hyper parameter setup. The convolution Layer of the CNN architecture process spatial, temporal, and spectral feature and generates the feature map in various context, generated feature map is max pooled in the pooling layer and classified into crops and medicinal Crop in the SoftMax layer. Experimental analysis of the proposed architecture is carried out on the Indiana Pines dataset using cross-fold validation to analyze the representation ability to discriminate the features with large variance between the different classes. From the results, it is confirmed that the proposed architecture exhibits higher performance in classification accuracy of 98.43% in classifying the Crop species compared with conventional approaches.
Article Details
References
M. Fauvel, J. A. Benediktsson, J. Chanussot, and J. R. Sveinsson, “Spectral and spatial classification of hyperspectral data using SVMs and morphological profiles,” IEEE Trans. Geosci. Remote Sens., vol. 46, no. 11, pp. 3804–3814, Nov. 2008
L. He, Y. Li, X. Li, and W. Wu, “Spectral–spatial classification of hyperspectral images via spatial translation-invariant wavelet-based sparse representation,” IEEE Trans. Geosci. Remote Sens., vol. 53, no. 5, pp. 2696– 2712, May 2015
P. Ghamisi et al., “New frontiers in spectral-spatial hyperspectral image classification: the latest advances based on mathematical morphology, Markov random fields, segmentation, sparse representation, and deep learning,” IEEE Geosci. Remote Sensing Mag., vol. 6, no. 3, pp. 10–43, Sep. 2018.
B. Waske, S. van der Linden, J. Benediktsson, A. Rabe, and P. Hostert, "Sensitivity of support vector machines to random feature selection in the classification of hyperspectral data," IEEE Trans. Geosci. Remote Sens., vol. 48, no. 7, pp. 2880–2889, Jul. 2010
J. Muñoz-Marí, F. Bovolo, L. Gómez-Chova, L. Bruzzone, and G. Camp-Valls, “Semisupervised one-class support vector machines for the classification of remote sensing data," IEEE Trans. Geosci. Remote Sens., vol. 48, no. 8, pp. 3188–3197, 2010.
J. Xia, M. D. Mura, J. Chanussot, P. Du, and X. He, “Random subspace ensembles for hyperspectral image classification with extended morphological attribute profiles,” IEEE Trans. Geosci. Remote Sens., vol. 53, no. 9, pp. 4768–4786, Sep. 2015.
Y. Y. Tang, H. Yuan, and L. Li, “Manifold-based sparse representation for hyperspectral image classification,” IEEE Trans. Geosci. Remote Sens., vol. 52, no. 12, pp. 7606–7618, Dec. 2014
P. Du, J. Xia, W. Zhang, K. Tan, Y. Liu, and S. Liu, "Multiple classifier systems for remote sensing image classification: A review," Sensors, vol. 12, no. 4, pp. 4764–4792, 2012.
J. Xia, P. Ghamisi, N. Yokoya, and A. Iwasaki, “Random forest ensembles and extended multi-extinction profiles for hyperspectral image classification,” IEEE Trans. Geosci. Remote Sens., vol. 56, no. 1, pp. 202–216, Jan. 2018.
B. Rivard, J. Feng, A. Gallie, and A. Sanchez-Azofeifa, “Continuous wavelets for the improved use of spectral libraries and hyperspectral data,” Remote Sens. Environ., vol. 112, pp. 2850–2862, 2008.
Schmalz, M.S., Ritter, G.X., Urcid, G.: Autonomous single-pass endmember approximation using lattice auto-associative memories. In: 10th Joint Conference on Information Sciences. Elsevier, Amsterdam (preprint, 2008) (Special Issue)
O. E. Malahlela, M. A. Cho, and O. Mutanga, “Mapping the occurrence of chromolaenaodorata (L.) in subtropical forest gaps using environmental and remote sensing data,” Biol. Invasions, vol. 17, pp. 2027–2042, Jul. 2015.
Ravi, G. ., Das, M. S. ., & Karmakonda, K. . (2023). Energy Efficient Data Aggregation Scheme using Improved LEACH Algorithm for IoT Networks. International Journal of Intelligent Systems and Applications in Engineering, 11(2s), 174 –. Retrieved from https://ijisae.org/index.php/IJISAE/article/view/2521
Y. Qian, M. Ye, and J. Zhou, “Hyperspectral image classification based on structured sparse logistic regression and three-dimensional wavelet texture features,” IEEE Trans. Geosci. Remote Sens., vol. 51, no. 4, pp. 2276–2291, Apr. 2013.
Y. Tarabalka, M. Fauvel, J. Chanussot, and J. A. Benediktsson, “SVMand MRF-based method for accurate classification of hyperspectral images,” IEEE Geosci. Remote Sens. Lett., vol. 7, no. 4, pp. 736–740, Oct. 2010.
K. Jia, B. Wu, Y. Tian, Y. Zeng, and Q. Li, “Vegetation classification method with biochemical composition estimated from remote sensing data,” Int. J. Remote Sens., vol. 32, pp. 9307–9325, 20
N. Thamaraikannan and S. Manju, "Review on Image Classification Techniques in Machine Learning for Satellite Imagery," 2021 International Conference on Artificial Intelligence and Smart Systems (ICAIS), Coimbatore, India, 2021, pp. 144-149, doi: 10.1109/ICAIS50930.2021.9395808.
N. Thamaraikannan and S. Manju," Classification of Satellite Agriculture Imagery to Identify Crop Type Using Deep Learning Techniques - A Review," 2023 International Conference on Computer Communication and Informatics (ICCCI), Jan. 23 – 25, 2023, Coimbatore, INDIA, 2023, 979-8-3503-4821-7/23/$31.00©2023 IEEE.
S Manju and N Thamaraikannan, "Predictive Analysis on Satellite Imagery Techniques", Journal of Shanghai Jiaotong University, vol. 16, no. 9, September 2020.
N. Thamaraikannan and S. Manju," Analysis of Support Vector Machine and Convolutional Neural Network to Improve Satellite Imagery," Indian Journal of Natural Sciences, Vol.12, Issue 70, pg.no38885- 38897),0976–0997.
Thamaraikannan, N., & Manju, S” A DETAILED ANALYSIS ON CLASSIFICATION AND FEATURE EXTRACTION TECHNIQUES FOR HYPER SPECTRAL REMOTE SENSING CROP IMAGES”, Eur. Chem. Bull. 2023, 12 (Special Issue 6), 4557– 4578, doi: 10.31838/ecb/2023.12.si6.4022023.08/06/2023