Residual Edge Attention in U-Net for Brain Tumour Segmentation
Main Article Content
Abstract
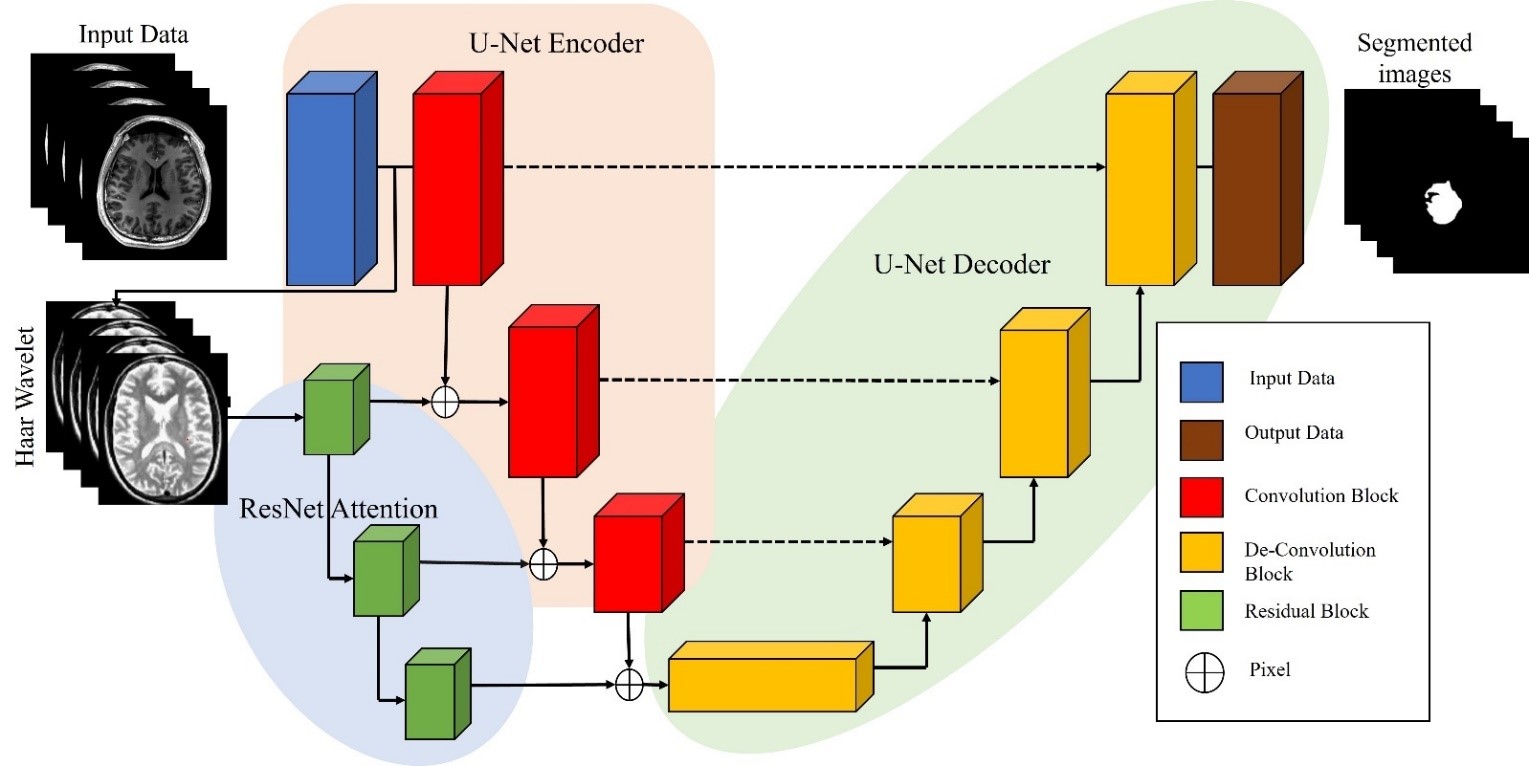
Identification and delineation of the tumour area in images of the brain constitute the crucial job of brain tumour segmentation in medical imaging. This task is crucial for diagnosis, treatment organizing, and keeping a track of brain tumours. Medical imaging methods like magnetic resonance imaging (MRI) or computed tomography scans are frequently used to divide brain tumours in real time. (CT). These imaging techniques provides high-resolution images for the brain that allows doctor to identify and locate tumours. There are several approaches to brain tumour segmentation, including manual segmentation by a radiologist, semi-automated segmentation using software tools that require some manual intervention, and fully automated segmentation using artificial intelligence (AI) algorithms. In this probing work, For segmenting brain tumours, we had anticipated Residual Edge Attention in U-Net design (ResEA-U-Net). Residual Edge Attention (ResEA) is a novel approach that enhances the performance of the U-Net architecture for brain tumour segmentation. The U-Net is often used in deep learning architecture for medical MRI brain image segmentation tasks, but it suffers from limited receptive field and feature reuse. To address this limitation, ResEA is expected to capture wide-range dependencies and enable network to focus on important regions of the image. The ResEA block contains of a residual block and an attention block that are connected in series. The residual block is created to improve the gradient flow and feature reuse, while the attention block focuses on important regions of the image by assigning higher weights to informative edges. The expected approach to evaluated on the BraTS data, which contain images of magnetic resonance of brain tumours. Experimental outcomes demonstrate that the ResEA-U-Net outperforms the baseline U-Net and other state-of-the-art methods. Overall, the suggested ResEA-U-Net architecture is a promising approach for brain tumour segmentation because it improves segmentation accuracy and lowers segmentation false positive rate, which can be essential for precise detection and therapy planning.
Article Details
References
Z. Liu et al., “Deep Learning Based Brain Tumour egmentation: A Survey,” vol. 14, no. 8, pp. 1–21, 2020, [Online]. Available: http://arxiv.org/abs/2007.09479.
A. P. Patel et al., “Global, regional, and national burden of brain and other CNS cancer, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016,” Lancet Neurol., vol.18, no. 4, pp. 376–393, 2019, doi: 10.1016/S1474-4422(18)30468-X.
M. Ghaffari, A. Sowmya, and R. Oliver, “Automated Brain Tumour Segmentation Using Cascaded 3D Densely Connected U-Net,” Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics), vol. 12658 LNCS, pp. 481–491, 2021, doi: 10.1007/978-3-030-72084-1_43.
E.-E. Mohd. Azhari, M. M. Mohd. Hatta, Z. Zaw Htike, and S. Lei Win, “Tumour detection in medical imaging: a Survey,” Int. J. Adv. Inf. Technol., vol. 4, no. 1, pp. 21–30, 2014, doi:10.5121/ijait.2013.4103.
D. N. Louis et al., “The 2016 World Health Organization Classification of Tumours of the Central Nervous System: a summary,” ActaNeuropathol., vol. 131, no. 6, pp. 803–820, 2016, doi:10.1007/s00401-016-1545-1.
S. A. L. I. R. Al-qazzaz, “Deep Learning-based Brain Tumour Image Segmentation and its Extension to Stroke Lesion Segmentation,” 2020.
R. V. Tanneedi, P. Pedapati, and S. Johansson, “Brain tumour detection using HOG by SVM,” no. December 2017, [Online]. Available: www.bth.se.
S. Bauer, R. Wiest, L. P. Nolte, and M. Reyes, “A survey of MRI-based medical image analysis for brain tumour studies,” Phys. Med. Biol., vol. 58, no. 13, 2013, doi: 10.1088/0031-9155/58/13/R97.
U. Baid et al., “A Novel Approach for Fully Automatic Intra-Tumour Segmentation With 3D U-Net Architecture for Gliomas,” Front. Comput. Neurosci., vol. 14, no. February, pp. 1–11, 2020, doi:10.3389/fncom.2020.00010.
W. Weng and X. Zhu, “INet: Convolutional Networks for Biomedical Image Segmentation,” IEEE Access, vol. 9, pp. 16591–16603, 2021, doi: 10.1109/ACCESS.2021.3053408.
A. Crimi et al., Glioma, Multiple Sclerosis , Stroke and Traumatic Brain Injuries, vol. 2. Springer International Publishing, 2018.
W. Wang, C. Chen, M. Ding, H. Yu, S. Zha, and J. Li, “TransBTS: Multimodal Brain Tumour Segmentation Using Transformer,” Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics), vol. 12901 LNCS, pp. 109–119, 2021, doi: 10.1007/978-3-030-87193-2_11.
P. Liu, Q. Dou, Q. Wang, and P. A. Heng, “An encoder-decoder neural network with 3D squeeze and-excitation and deep supervision for brain tumour segmentation,” IEEE Access, vol. 8, pp.34029–34037, 2020, doi: 10.1109/ACCESS.2020.2973707.
Z. Huang, Y. Zhao, Y. Liu, and G. Song, “GCAUNet: A group cross-channel attention residual UNet for slice-based brain tumour segmentation,” Biomed. Signal Process. Control, vol. 70, no. June, p. 102958, 2021, doi: 10.1016/j.bspc.2021.102958.
J. Zhang, X. Lv, H. Zhang, and B. Liu, “AResU-Net: Attention residual U-Net for brain tumour segmentation,” Symmetry (Basel)., vol. 12, no. 5, pp. 1–15, 2020, doi: 10.3390/SYM12050721.
D. Maji, P. Sigedar, and M. Singh, “Attention Res-UNet with Guided Decoder for semantic segmentation of brain tumours,” Biomed. Signal Process. Control, vol. 71, no. PA, p. 103077, 2022, doi: 10.1016/j.bspc.2021.103077.
M. K. Abd-Ellah, A. A. M. Khalaf, A. I. Awad, and H. F. A. Hamed, TPUAR-net: two parallel Unet with asymmetric residual-based deep convolutional neural network for brain tumour segmentation, vol. 11663 LNCS. Springer International Publishing, 2019.
J. Zhang, Z. Jiang, J. Dong, Y. Hou, and B. Liu, “Attention Gate ResU-Net for Automatic MRI Brain Tumour Segmentation,” IEEE Access, vol. 8, pp. 58533–58545, 2020, doi: 10.1109/ACCESS.2020.2983075.
S. Qamar, P. Ahmad, and L. Shen, “HI-Net: Hyperdense Inception 3D UNet for Brain Tumour Segmentation,” Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics), vol. 12659 LNCS, pp. 50–57, 2021, doi: 10.1007/978-3-030-72087-2_5.
F. Isensee, P. F. Jäger, P. M. Full, P. Vollmuth, and K. H. Maier-Hein, “nnU-Net for Brain Tumour Segmentation,” Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics), vol. 12659 LNCS, pp. 118–132, 2021, doi: 10.1007/978-3-030-72087-2_11.
M. Havaei et al., “Brain tumour segmentation with Deep Neural Networks,” Med. Image Anal., vol. 35, pp. 18–31, 2017, doi: 10.1016/j.media.2016.05.004.
M. I. Razzak, M. Imran, and G. Xu, “Efficient Brain Tumour Segmentation with Multiscale Two Pathway-Group Conventional Neural Networks,” IEEE J. Biomed. Heal. Informatics, vol. 23, no.5, pp. 1911–1919, 2019, doi: 10.1109/JBHI.2018.2874033.
M. Yaqub et al., “State-of-the-art CNN optimizer for brain tumour segmentation in magnetic resonance images,” Brain Sci., vol. 10, no. 7, pp. 1–19, 2020, doi: 10.3390/brainsci10070427.
B. H. Menze et al., “The Multimodal Brain Tumour Image Segmentation Benchmark (BRATS),” IEEE Trans. Med. Imaging, vol. 34, no. 10, pp. 1993–2024, 2015, doi:10.1109/TMI.2014.2377694.
K. He, X. Zhang, S. Ren, and J. Sun, “Deep residual learning for image recognition,” Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit., vol. 2016-Decem, pp. 770–778, 2016, doi:10.1109/CVPR.2016.90.
Ö. Çiçek, A. Abdulkadir, S. S. Lienkamp, T. Brox, and O. Ronneberger, “3D U-net: Learning dense volumetric segmentation from sparse annotation,” Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics), vol. 9901 LNCS, pp. 424–432, 2016, doi: 10.1007/978-3-319-46723-8_49.
S. Ioffe and C. Szegedy, “Batch normalization: Accelerating deep network training by reducing internal covariate shift,” 32nd Int. Conf. Mach. Learn. ICML 2015, vol. 1, pp. 448–456, 2015.
L. H. Shehab, O. M. Fahmy, S. M. Gasser, and M. S. El-Mahallawy, “An efficient brain tumour image segmentation based on deep residual networks (ResNets),” J. King Saud Univ. - Eng. Sci.,vol. 33, no. 6, pp. 404–412, 2021, doi: 10.1016/j.jksues.2020.06.001.
C. H. Sudre, W. Li, T. Vercauteren, S. Ourselin, and M. Jorge Cardoso, “Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations,” Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics), vol. 10553 LNCS, pp.240–248, 2017, doi: 10.1007/978-3-319-67558-9_28.
F. Milletari, N. Navab, and S. A. Ahmadi, “V-Net: Fully convolutional neural networks for volumetric medical image segmentation,” Proc. - 2016 4th Int. Conf. 3D Vision, 3DV 2016, pp.565–571, 2016, doi: 10.1109/3DV.2016.79.
T. Y. Lin, P. Goyal, R. Girshick, K. He, and P. Dollar, “Focal Loss for Dense Object Detection,” Proc. IEEE Int. Conf. Comput. Vis., vol. 2017-Octob, pp. 2999–3007, 2017, doi:10.1109/ICCV.2017.324.
J. Colman, L. Zhang, W. Duan, and X. Ye, “DR-Unet104 for Multimodal MRI Brain Tumour Segmentation,” Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics), vol. 12659 LNCS, no. 2021, pp. 410–419, 2021, doi: 10.1007/978-3-030-72087-2_36.
L. M. Ballestar and V. Vilaplana, “Brain Tumour Segmentation using 3D-CNNs with Uncertainty Estimation,” pp. 1–11, 2020, [Online]. Available: http://arxiv.org/abs/2009.12188.
H. Messaoudi et al., “Efficient Embedding Network for 3D Brain Tumour Segmentation,” Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics), vol.12658 LNCS, pp. 252–262, 2021, doi: 10.1007/978-3-030-72084-1_23.
F. Wang, R. Jiang, L. Zheng, C. Meng, and B. Biswal, “3D U-Net Based Brain Tumour Segmentation and Survival Days Prediction,” Lect. Notes Comput. Sci. (including Subser. Lect.Notes Artif. Intell. Lect. Notes Bioinformatics), vol. 11992 LNCS, pp. 131–141, 2020, doi:10.1007/978-3-030-46640-4_13.
A. Kleppe, O. J. Skrede, S. De Raedt, K. Liestøl, D. J. Kerr, and H. E. Danielsen, “Designing deep learning studies in cancer diagnostics,” Nat. Rev. Cancer, vol. 21, no. 3, pp. 199–211, 2021, doi:10.1038/s41568-020-00327-9.
Messaoudi, H., Belaid, A., Allaoui, M.L., Zetout, A., Allili, M.S., Tliba, S., Salem, D.B., and Conze, P.-H.: ‘Efficient embedding network for 3D brain tumour segmentation’, arXiv preprintarXiv:2011.11052, 2020.
R. R. Agravat and M. S. Raval, “3d semantic segmentation of brain tumour for overall survival prediction,” in International MICCAI Brainlesion Workshop. Springer, 2020, pp. 215–227.
V. K. Anand, S. Grampurohit, P. Aurangabadkar, A. Kori, M. Khened, R. S. Bhat, and G. Krishnamurthi, “Brain tumour segmentation and survival prediction using automatic hard mining in 3d cnn architecture,” in International MICCAI Brainlesion Workshop. Springer, 2020, pp. 310–319.
Haozhe Jia 1 , 2 , 4 , Weidong Cai 3 , Heng Huang 4 , 5 , and Yong Xia1,2 .1 Research & Development Institute of Northwestern Polytechnical University in Shenzhen, Shenzhen 518057, China yxia@nwpu.edu.cn 2 National Engineering Laboratory for Integrated Aero-Space-Ground-Ocean Big Data Application Technology, School of Computer Science and Engineering, Northwestern Polytechnical University, Xi’an 710072, China 3 School of Computer Science, University of Sydney, Sydney, NSW 2006, Australia 4 Department of Electrical and Computer Engineering, University of Pittsburgh, Pittsburgh, PA 15261, USA 5 JD Finance America Corporation, California, CA 94043, US
Theophraste Henry 1 *, Alexandre Carre 1 *, Marvin Lerousseau 1, 2, Theo Estienne 1,2 , Charlotte Robert 1 , 3 , Nikos Paragios 4 , and Eric Deutsch 1 , 3 1 University’s Paris-Saclay, Institute Gustave Roussy, Inserm, Radioth´erapie Mol´eculaire et Innovation Therapeutae, F-94805, Villejuif, France. 2 University Paris-Saclay, CentraleSuplec, 91190, Gif-sur-Yvette, France. 3 Gustave Roussy, Department d’oncologie-radioth´erapie, F-94805, Villejuif, France 4 Thera panacea, Paris, France
Carlos A. Silva 1 , Adriano Pinto 1 , S´ergio Pereira 1 , and Ana Lopes 1 Center MEMS of University of Minho, Campus of Azur´em, 4800-058 Guimar˜aes Portugal csilva@dei.uminho.pt
Yixin Wang 1 , 2 , 3 , Yao Zhang 1 , 2 , 3 , Feng Hou 1 , 2 , 3 , Yang Liu 1 , 2 , 3 , Jiang Tian 3 , Cheng Zhong 3 , Yang Zhang 4 , and Zhiqiang He 1 , 2 , 4 1 Institute of Computing Technology, Chinese Academy of Sciences, Beijing, China 2 University of Chinese Academy of Sciences, Beijing, China. 3 AI Lab, Lenovo Research, Beijing, China. Lenovo Corporate Research & Development, Lenovo Ltd., Beijing, China.
V.K.Anand, S.Grampurohit, P. Aurangabadkar, A. Kori, M. Khened, R. S. Bhat, and G. Krishnamurthi, “Brain tumour segmentation and survival prediction using automatic hard mining in 3d cnn architecture,” in International MICCAI Brainlesion Workshop. Springer, 2020, pp. 310–319.
Yu Liu , Member, IEEE, Fuhao Mu, Yu Shi , and Xun Chen , Senior Member, IEEE, “SF-Net: A Multi-Task Model for Brain Tumour Segmentation in Multimodal MRI via Image Fusion “ .
MD Abdullah Al Nasim? , Abdullah Al Munem? , Maksuda Islam? , Md Aminul Haque Palash? , MD. Mahim Anjum Haque? , and Faisal Muhammad Shah† ?Dept of Research and Development, Pioneer Alpha †Dept of Computer Science and Engineering, AUST
Yading Yuan Department of Radiation Oncology Icahn School of Medicine at Mount Sinai New York, NY, USA
[48] JIANXIN ZHANG 1,2, (Member, IEEE), ZONGKANG JIANG1 , JING DONG1 , YAQING HOU 3 , AND BIN LIU4,5 “Attention Gate ResU-Net for Automatic MRI Brain Tumour Segmentation”.
M. Hamghalam, B. Lei, and T. Wang, “Brain tumour synthetic segmentation in 3D multimodal MRI scans,” in International MICCAI Brainlesion Workshop, 2019, pp. 153–162.
Z. Jiang, C. Ding, M. Liu, and D. Tao, “Two-Stage Cascaded U-Net: 1st Place Solution to BraTS Challenge 2019 Segmentation Task,” in International MICCAI Brainlesion Workshop, 2019, pp. 231–241 Z. Jiang, C. Ding, M. Liu, and D. Tao, “Two-Stage Cascaded U-Net: 1st Place Solution to BraTS Challenge 2019 SegmentationTask,” in International MICCAI Brainlesion Workshop, 2019, pp. 231–241
M. Frey and M. Nau, “Memory efficient brain tumour segmentation using an autoencoder-regularised U-Net,” inInternational MICCAI Brainlesion Workshop, 2019, pp. 388–396.
Doyle, S.; Vasseur, F.; Dojat, M.; Forbes, F.: Fully automatic brain tumour segmentation from multiple MR sequences using hidden Markov felds and variational EM. Proc. NCI MICCAI BRATS 1,18–22 (2013)
Zhao, X., Wu, Y., Song, G., Li, Z., Zhang, Y., & Fan, Y. (2018). A deep learning model integrating FCNNs and CRFs for brain tumour segmentation. Medical Image Analysis, 43, 98–111.
Havaei, M. et al. (2017). Brain tumour segmentation with deep neural networks. Medical Image Analysis, 35, 18–31.
ikic, D., Ioannou, Y., Brown, M., & Criminisi, A. (2014). Segmentation of brain tumour tissues with convolutional neural networks. Proceedings MICCAI-BRATS, 36–39.
Dong, H., Yang, G., Liu, F., Mo, Y., & Guo, Y. (2017). Automatic braintumour detection and segmentation using U-Net based fully convolutional networks. In Annual conference on medical image understanding
Dong, H., Yang, G., Liu, F., Mo, Y., & Guo, Y. (2017). Automatic braintumour detection and segmentation using U-Net based fully convolutional networks. In Annual conference on medical image understanding
Kamnitsas, K. et al. (2017). Efcient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Medical Image Analysis, 36, 61–78.
Chenhong Zhou, Changxing Ding , Member, IEEE, Xinchao Wang , Member, IEEE, Zhentai Lu, Member, IEEE, and Dacheng Tao , Fellow, IEEE
Fabian Isensee1 , Philipp Kickingereder2 , Wolfgang Wick3 , Martin Bendszus2 , and Klaus H. Maier-Hein1 1 Division of Medical Image Computing, German Cancer Research Center (DKFZ), Heidelberg, Germany 2 Department of Neuroradiology, Heidelberg University Hospital, Heidelberg, Germany 3 Neurology Clinic, Heidelberg University Hospital, Heidelberg, Germany
Yi Ding , Member, IEEE, Wei Zheng, Ji Geng, Zhen Qin , Member, IEEE, Kim-Kwang Raymond Choo , Senior Member, IEEE, Zhiguang Qin , Member, IEEE, and Xiaolin Hou.” MVFusFra: A Multi-View Dynamic Fusion Framework for Multimodal Brain Tumour Segmentation”.
KAI HU 1,2, (Member, IEEE), QINGHAI GAN1 , YUAN ZHANG1 , SHUHUA DENG 1 , FEN XIAO1 , WEI HUANG4 , CHUNHONG CAO 1 , AND XIEPING GAO 1,3, (Member, IEEE) “Brain Tumour Segmentation Using Multi-Cascaded Convolutional Neural Networks and Conditional Random Field”.
Pereira S, Pinto A, Alves V, Silva C A. Brain tumour segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging 2016;35(5):1240–1251.
Dvorak P, Menze B. Structured prediction with convolutional neural networks for multimodal brain tumour segmentation. MICCAI Multimodal Brain Tumour Segmentation Challenge (BraTS) 2015:13–24.
Mohammad Havaei ?1 , Hugo Larochelle†1,2, Philippe Poulin‡1 , and Pierre-Marc Jodoin§1,3 1Universit´e de Sherbrooke, Sherbrooke, Canada 2Twitter, USA 3 Imeka, Canada.” Within-Brain Classification for Brain Tumour Segmentation”.
NEIL MICALLEF 1 , (Member, IEEE), DYLAN SEYCHELL 1 , (Senior Member, IEEE), AND CLAUDE J. BAJADA “Exploring the U-Net++ Model for Automatic Brain Tumour Segmentation”.
N. J. Tustison et al., ‘‘Optimal symmetric multimodal templates and concatenated random forests for supervised brain tumour segmentation (simplified) with ANTsR,’’ Neuroinformatics, vol. 13, no. 2, pp. 209–225, 2015
2. Hu K., Deng S.H. Brain Tumour Segmentation Using Multi-Cascaded Convolutional Neural Networks and Conditional Random Field. IEEE Access. 2019;7:2615–2629. doi: 10.1109/ACCESS.2019.2927433.
Bhagat P.K., Choudhary P. Multi-class Segmentation of Brain Tumour from MRI Images. Appl. Artif. Intell. Tech. Eng. Adv. Intell. Syst. Comput. 2019;698:543–553.
Ieva A., Russo C., Liu S., Jian A., Bai M.Y., Qian Y., Magnussen J.S. Application of Deep Learning for Automatic Segmentation of Brain Tumours on Magnetic Resonance Imaging: A Heuristic Approach in the Clinical Scenario. Neuroradiology. 2021;63:1253–1262. doi: 10.1007/s00234-021-02649-3