Efficient Disease Identification Method for Crop Leaf using Deep Learning Techniques
Main Article Content
Abstract
Many prime grain-producing nations have implemented steps to limit export of grains as COVID-19 has expanded over the globe; food security has sparked significant worry from a number of stakeholders. One of the most crucial concerns facing all nations is how to increase grain output. However, the diseases occur in crops remain a challenge for countless farmers, therefore it is critical to understand their severity promptly and precisely to guide the them in taking additional measures to lessen the chances of plants being affected furthermore.
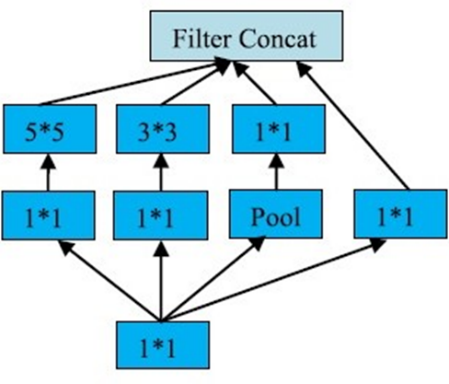
This paper describes a deep learning model for the identification of crop diseases that can achieve high accuracy with low processing power. The model, called the inception v3 network, has been tested on a tomato leaf dataset and has obtained a average identification accuracy of 98.00% and further the ensemble of two inception v3 models with slight diversity achieved an accuracy of 98.11%. The results suggest that this model could be useful in improving food security by helping farmers quickly and accurately identify crop diseases and take appropriate measures to prevent further spread.
Article Details
References
(Oct. 2020). Mitigating Impacts of COVID-19 on Food Trade and Mar- kets. [Online]. Available: http://www.fao.org/news/story/en/item/1268719/ icode
B. Keswani, A. G. Mohapatra, A. Mohanty, A. Khanna,J. J. P. C. Rodrigues, D. Gupta, and V. H. C. de Albuquerque, “Adapting weather conditions based IoT enabled smart irrigation technique in precision agriculture mechanisms,” Neural Comput. Appl.,, vol. 31, no. S1, pp. 277- 292, Jan. 2019,doi: 10.1007/s00521-018-3737-1.
B. Keswani, G. A. Mohapatra, P. Keswani, A. Khanna, D. Gupta, and J. Rodrigues, “Improving weather dependent zone specific irrigation control scheme in IoT and big data enabled self driven precision agriculture mechanism,” Enterprise Inf. Syst.,, vol. 14, pp. 9-10, 1494-1515, 2020, doi: 10.1080/17517575.2020.1713406.
D. Jiang, F. Li, Y. Yang, and S. Yu, “A tomato leaf diseases classifi- cation method based on deep learning,” in Proc. Chin. Control Decis. Conf. (CCDC),,Aug. 2020, pp. 1446-1450, doi: 10.1109/CCDC49329. 2020.9164457.
P. Sharma, P. Hans, and S. C. Gupta, “Classification of plant leaf diseases using machine learning and image preprocessing techniques,” in Proc. 10th Int. Conf. Cloud Comput., Data Sci. Eng. (Confluence),,Jan. 2020, pp. 480- 484, doi: 10.1109/Confluence47617.2020.9057889.
M. Lv, G. Zhou, M. He, A. Chen, W. Zhang, and Y. Hu, “Maize leaf disease identification based on feature enhancement and DMS-robust alexnet,” IEEE Access,, vol. 8, pp. 57952-57966, 2020, doi: 10.1109/AC- CESS.2020.2982443.
B. Liu, C. Tan, S. Li, J. He, and H. Wang, “A data augmentation method based on generative adversarial networks for grape leaf dis- ease identification,” IEEE Access,, vol. 8, pp. 102188-102198, 2020, doi: 10.1109/ACCESS.2020.2998839.
S. Liang and W. Zhang, “Accurate image recognition of plant diseases based on multiple classifiers integration,”, in Proc. Chin. Intell. Syst. Conf.,, vol. 594, 2020, pp. 103-113, doi: 10.1007/978-981-32-9698-5-13.
S. M. Jaisakthi, P. Mirunalini, D. Thenmozhi, and Vatsala, “Grape leaf disease identification using machine learning techniques,” in Proc. Int. Conf. Comput. Intell. Data Sci. (ICCIDS),, Feb. 2019, pp. 1-6, doi: 10.1109/ ICCIDS.2019.8862084.
S. Huang, W. Liu, F. Qi, and K. Yang, “Development and valida- tion of a deep learning algorithm for the recognition of plant dis- ease,” in Proc. IEEE 21st Int. Conf. High Perform. Comput. Commun., IEEE 17th Int. Conf. Smart City, IEEE 5th Int. Conf. Data Sci. Syst.(HPCC/SmartCity/DSS),,Aug. 2019, pp. 1951-1957, doi: 10.1109/HPCC/SmartCity/DSS.2019.00269.
A. Waheed, M. Goyal, D. Gupta, A. Khanna, A. E. Hassanien, and H. M. Pandey, “An optimized dense convolutional neural network model for disease recognition and classification in corn leaf,” Comput. Elec- tron. Agricult.,, vol. 175, Aug. 2020, Art. no. 105456, doi: 10.1016/ j.compag.2020.105456.
Q. Tian, J. Li, and H. Liu, “A method for guaranteeing wireless communication based on a combination of deep and shallowlearn- ing,” IEEE Access,,vol. 7, pp. 38688-38695, 2019, doi: 10.1109/AC- CESS.2019.2905754.
B. M. Aslahi-Shahri, R. Rahmani, M. Chizari, A. Maralani, M. Eslami, M. J. Golkar, and A. Ebrahimi,“A hybrid method consisting of GA and SVM for intrusion detection system,” Neural Comput. Appl.,,vol. 27, no. 6, pp. 1669-1676, Aug. 2016.
A.-C. Enache and V. Sgarciu, “Anomaly intrusions detection based on support vector machines with an improved bat algorithm,” in Proc. 20th Int. Conf. Control Syst. Comput. Sci.,,May 2015, pp. 317-321.
A.-C. Enache and V. V. Patriciu, “Intrusions detection based on sup- port vector machine optimized with swarm intelligence,” in Proc. IEEE 9th IEEE Int. Symp. Appl. Comput. Intell. Informat. (SACI),,May 2014, pp. 153-158.
N. R. Bhimte and V. R. Thool, “Diseases detection of cotton leaf spot using image processing and SVM classifier,” in Proc. 2nd Int. Conf. Intell. Comput. Control Syst. (ICICCS),,Jun. 2018, pp. 340-344, doi: 10.1109/ ICCONS.2018.8662906.
Prof. Muhamad Angriawan. (2016). Performance Analysis and Resource Allocation in MIMO-OFDM Systems. International Journal of New Practices in Management and Engineering, 5(02), 01 - 07. Retrieved from http://ijnpme.org/index.php/IJNPME/article/view/44.
P. B. Padol and A. A. Yadav, “SVM classifier based grape leaf dis- ease detection,” in Proc. Conf. Adv. Signal Process. (CASP),,Jun. 2016, pp. 175-179, doi: 10.1109/CASP.2016.7746160.
O. Russakovsky, J. Deng, H. Su, J. Krause, S. Satheesh, S. Ma, Z. Huang, A. Karpathy, A. Khosla, M. Bernstein, A. C. Berg, and L. Fei-Fei, “ImageNet large scale visual recognition challenge,” Int. J. Comput. Vis., vol. 115, no. 3, pp. 211–252, Dec. 2015.
F. Konidaris, T. Tagaris, M. Sdraka, and A. Stafylopatis, “Generative adversarial networks as an advanced data augmentation technique for MRI data,” in Proc. 14th Int. Conf. Comput. Vis. Theor. Appl., Jan. 2019, pp. 48–59, doi: 10.5220/0007363900480059.
M. Alhawarat and M. Hegazi, “Revisiting K-means and topic modeling, a comparison study to cluster arabic documents,”IEEE Access,,vol. 6,pp. 42740-42749, 2018.
Y. Meng, J. Liang, F. Cao, and Y. He, “A new distance with deriva- tive information for functional k-means clustering algorithm,”Inf. Sci.,,vols. 463-464, pp. 166-185, Oct. 2018.
Z. Lv, T. Liu, C. Shi, J. A. Benediktsson, and H. Du, “Novel land cover change detection method based on k-Means clustering and adaptive major- ity voting using bitemporal remote sensing images,”IEEE Access,,vol. 7, pp. 34425-34437, 2019.
J. Zhu, Z. Jiang, G. D. Evangelidis, C. Zhang, S. Pang, and Z. Li, “Efficient registration of multi-view point sets by K-means clustering,”Inf. Sci.,vol. 488, pp. 205-218, Jul. 2019.
S. Zhang, H.Wang,W. Huang, and Z. You, “Plant diseased leaf segmentation and recognition by fusion of superpixel, K- means and PHOG,”Optik,vol. 157, pp. 866-872, Mar. 2018, doi: 10.1016/j.ijleo.2017.11.190.
Hee Kim, M. . (2023). Exploring the Singularity Between Google Searches and Suicide Deaths After Celebrity Suicides. International Journal of Intelligent Systems and Applications in Engineering, 11(4s), 180–187. Retrieved from https://ijisae.org/index.php/IJISAE/article/view/2586.
R. Anand, S. Veni, and J. Aravinth, “An application of image pro- cessing techniques for detection of diseases on brinjal leaves using k- means clus- tering method,”in Proc. Int. Conf. Recent Trends Inf. Technol. (ICRTIT),Apr. 2016, pp. 1-6, doi: 10.1109/ICRTIT.2016.7569531.
C. U. Kumari, S. Jeevan Prasad, and G. Mounika, “Leaf disease detection: Feature extraction with K-means clustering and classification with ANN,”in Proc. 3rd Int. Conf. Comput. Methodologies Commun. (ICCMC),Mar. 2019, pp. 1095-1098, doi: 10.1109/ICCMC.2019.8819750.
F. A. P. Rani, S. N. Kumar, A. L. Fred, C. Dyson, V. Suresh, and P. S. Jeba, “K-means clustering and SVM for plant leaf disease detec- tion and classification,”in Proc. Int. Conf. Recent Adv. Energy- Efficient Comput. Commun. (ICRAECC),Mar. 2019, pp. 1-4, doi: 10.1109/ ICRAECC43874.2019.8995157.
A. Krizhevsky, “One weird trick for parallelizing convolutional neural networks,”2014, arXiv:1404.5997. [Online]. Available: http://arxiv.org/abs/1404.5997
Q. Zeng, X. Ma, B. Cheng, E. Zhou, and W. Pang, “GANs-based data augmentation for citrus disease severity detection using deep learn- ing,”IEEE Access,vol. 8, pp. 172882-172891, 2020, doi: 10.1109/AC- CESS.2020.3025196.
Juan Garcia, Guðmundsdóttir Anna, Maria Jansen, Johansson Anna, Anna Wagner. Exploring Decision Trees and Random Forests for Decision Science Applications. Kuwait Journal of Machine Learning, 2(4). Retrieved from http://kuwaitjournals.com/index.php/kjml/article/view/211.
C. Szegedy and Wei Liu and Yangqing Jia and P. Sermanet and S. Reed and D. Anguelov and D. Erhan and V. Vanhoucke and A. Rabinovich., "Going deeper with convolutions," in 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 2015 pp. 1-9.
doi: 10.1109/CVPR.2015.7298594
K. He, X. Zhang, S. Ren, and J. Sun, “Deep residual learning for image recognition,”in Proc. IEEE Conf. Comput. Vis. Pattern Recognit. (CVPR),Jun. 2016, pp. 770-778, doi: 10.1109/CVPR.2016.90.
G. Huang, Z. Liu, L. Van Der Maaten, and K. Q. Weinberger, “Densely connected convolutional networks,”in Proc. IEEE Conf. Com- put. Vis. Pattern Recognit. (CVPR),Jul. 2017, pp. 2261-2269, doi: 10.1109/CVPR.2017.243.
Y. Zhang, Y. Tian, Y. Kong, B. Zhong, and Y. Fu, “Resid- ual dense network for image super-resolution,”in Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recognit.,Jun. 2018, pp. 2472-2481, doi: 10.1109/CVPR.2018.00262.
D.-W. Kim, J. R. Chung, and S.-W. Jung, “GRDN: Grouped resid- ual dense network for real image denoising and GAN-based real- world noise modeling,”in Proc. IEEE/CVF Conf. Comput. Vis. Pat- tern Recognit. Workshops(CVPRW),Jun. 2019, pp. 2086-2094, doi: 10.1109/CVPRW.2019.00261.
C. Zhou, S. Zhou, J. Xing and J. Song, ”Tomato Leaf Disease Identi- fication by Restructured Deep Residual Dense Network,” IEEE Access, vol. 9, pp. 28822-28831, 2021, doi: 10.1109/ACCESS.2021.3058947.
B. Wang and D. Wang, “Plant leaves classification: A few-shot learning method based on siamese network,” IEEE Access, vol. 7, pp. 151754–151763, 2019.
S.-E.-A. Raza, G. Prince, J. P. Clarkson, and N. M. Rajpoot, “Automatic detection of diseased tomato plants using thermal and stereo visible light images,”PLoS ONE,vol. 10, no. 4, Apr. 2015, Art. no. e0123262.
S. Prasad, S. K. Peddoju, and D. Ghosh, “Multi-resolution mobile vision system for plant leaf disease diagnosis,”Signal, Image Video Process.,vol. 10, no. 2, pp. 379-388, Feb. 2016, doi: 10.1007/s11760-015-0751-y.
R. G. de Luna, E. P. Dadios, and A. A. Bandala, “Automated image capturing system for deep learning-based tomato plant leaf disease detec- tion and recognition,”in Proc. TENCON IEEE Region Conf.,Oct. 2018,pp. 1414-1419, doi: 10.1109/TENCON.2018.8650088.
X. Q. Guo, T. J. Fan, and X. Shu, “Tomato leaf diseases recognition based on improved multi-scale AlexNet,”Trans. Chin. Soc. Agr. Eng.,vol. 35, no. 13, pp. 162-169, Jul. 2019.
Q. Wu, Y. Chen, and J. Meng, “DCGAN-based data augmentation for tomato leaf disease identification,”IEEE Access,vol. 8, pp. 98716- 98728,2020, doi: 10.1109/ACCESS.2020.2997001.