Colorectal Cancer Classification from Protein Sequences Using Several RNN Pre-Trained Models
Main Article Content
Abstract
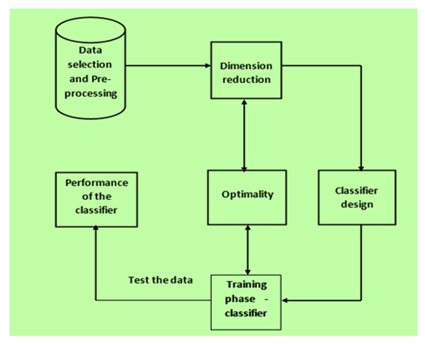
Bioinformatics is one field that can integrate health information with data mining applications in order to predict or analyze the patient’s information. One among the several diseases is colorectal cancer,which is horrible and rated the third leading disease in the area of cancer which leads to death in both men and women.In general, there are several intelligent methods to identify several kinds of problems in gene selection for predicting cancer,but there is no method to give a solution for patients who are diagnosed in the advanced stage.This motivated me to design theproposed approach in which we try to identify all the homo protein sequences and then train these sequences corresponding to one which causes colorectal cancer.InBioinformatics,a protein acts as one of the main agent or source to perform a biological function by interacting with molecules like Deoxyribonucleic acid (DNA), Ribonucleic acid (RNA). The function of a protein determines the healthy or diseased states of an organism. Protein interaction with other proteins can be visualized through a network called Protein-Protein Interaction Network (PPIN).In general classification of protein sequences is a very complex task. Deep learning techniques like CNN and RNN can be used to solve the problem.In computational bioinformatics,the classification of protein sequence plays an important role in determining accuracy.To improve the accuracy of our current model, the suggested method incorporates GRU, LSTM, RNN, and Customized LSTM into an RNN based architecture by optimizing the parameters in a two-way direction.Here we try to test all the models on sample protein sequences that are collected from TCGA and then determine the correctness of testing data and training data.
Article Details
References
Hatim Z Almarzouki, "Deep-Learning-Based Cancer Profiles Classification Using GeneExpressionData Profile", Journal of Healthcare Engineering, vol. 2022, Article ID 4715998, 2022. https://doi.org/10.1155/2022/4715998.
Sam Gelman, Sarah A. Fahlberg, Pete Heinzelman, Philip A. Romero, Anthony Gitter,”Neuralnetworks to learn protein sequence–function relationships from deep mutationalscanning data”,Proceedings of the National Academy of Sciences Nov 2021,118 (48)e2104878118;DOI: 10.1073/pnas.2104878118.
Tran KA, Kondrashova O, Bradley A, Williams ED, Pearson JV, Waddell N. Deep learningin cancerdiagnosis, prognosis and treatment selection. Genome Med. 2021;13(1):152.Published2021 Sep 27.doi:10.1186/s13073-021-00968-x.
Alzubaidi, L., Zhang, J., Humaidi, A.J. et al. Review of deep learning: concepts, CNNarchitectures,challenges,applications, future directions. J Big Data 8, 53 (2021).https://doi.org/10.1186/s40537-021-00444-8.
Unnam, A. K. ., & Rao, B. S. . (2023). An Extended Clusters Assessment Method with the Multi-Viewpoints for Effective Visualization of Data Partitions. International Journal of Intelligent Systems and Applications in Engineering, 11(1s), 51–56. Retrieved from https://ijisae.org/index.php/IJISAE/article/view/2476
Guo, L., Wang, S., Li, M. et al. Accurate classification of membrane protein types based onsequenceand evolutionary information using deep learning. BMC Bioinformatics 20, 700(2019).https://doi.org/10.1186/s12859-019-3275-6.
Prof. Sharayu Waghmare. (2012). Vedic Multiplier Implementation for High Speed Factorial Computation. International Journal of New Practices in Management and Engineering, 1(04), 01 - 06. Retrieved from http://ijnpme.org/index.php/IJNPME/article/view/8
Sofia EdströmJosefinOndrus,” Sequence Classification Applied To User Log Data,2016,SOFIA EDSTRÖM, JOSEFIN ONDRUS, June 2017”,Pp.9-12.
Anna, G., Hernandez, M., García, M., Fernández, M., & González, M. Optimizing Course Recommendations for Engineering Students Using Machine Learning. Kuwait Journal of Machine Learning, 1(1). Retrieved from http://kuwaitjournals.com/index.php/kjml/article/view/104
XingyouWang ,Weijie Jiang , Zhiyong Luo,” Combination Of Convolutional And RecurrentNeuralNetwork For Sentiment Analysis Of Short Texts”, Proceedings Of COLING 2016,The 26thInternationalConference On Computational Linguistics: Technical Papers, Osaka, Japan,December 11-17 2016, Pages 2428–2437.
Susan P. Imberman,” Effective Use of The Kdd Process and Data Mining for ComputerPerformanceProfessionals”:https://www.researchgate.net/ publication/ 221445402, NOV-2014.
Sofia Martinez, Machine Learning-based Fraud Detection in Financial Transactions , Machine Learning Applications Conference Proceedings, Vol 1 2021.
Muhammad Javed Iqbal, Ibrahima Faye, BrahimBelhaouari Samir, Abas MdSaid, "Efficient FeatureSelection and Classification of Protein Sequence Data inBioinformatics", The Scientific WorldJournal, vol. 2014, Article ID 173869, 12 pages,2014.https://doi.org/10.1155/2014/ 173869.
Khatri, K. ., & Sharma, D. A. . (2020). ECG Signal Analysis for Heart Disease Detection Based on Sensor Data Analysis with Signal Processing by Deep Learning Architectures. Research Journal of Computer Systems and Engineering, 1(1), 06–10. Retrieved from https://technicaljournals.org/RJCSE/index.php/journal/article/view/11
Hussein Hijazi1 and Christina Chan,A Classification Framework Applied to Cancer GeneExpressionProfiles,PMID: 23778014,doi: 10.1260/2040-2295.4.2.255,J Healthc Eng. 2013; 4(2): 10.1260/2040-2295.4.2.255.
John N. Weinstein, Eric A. Collisson, Gordon B. Mills, Kenna M. Shaw, Brad A.Ozenberger, KyleEllrott, Ilya Shmulevich, Chris Sander, Joshua M. Stuart,The CancerGenome Atlas Pan-CancerAnalysisProject,PMID: 24071849,Nat Genet. 2013Oct;45(10): pp1113–1120.doi: 10.1038/ng.2764