En-PaFlower: An Ensemble Approach using PSO and Flower Pollination Algorithm for Cancer Diagnosis
Main Article Content
Abstract
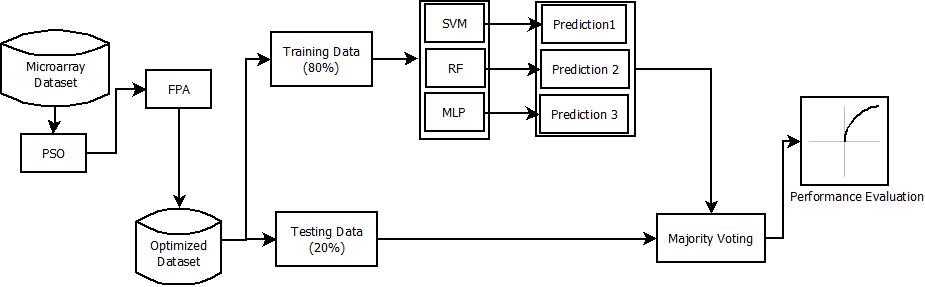
Machine learning now is used across many sectors and provides consistently precise predictions. The machine learning system is able to learn effectively because the training dataset contains examples of previously completed tasks. After learning how to process the necessary data, researchers have proven that machine learning algorithms can carry out the whole work autonomously. In recent years, cancer has become a major cause of the worldwide increase in mortality. Therefore, early detection of cancer improves the chance of a complete recovery, and Machine Learning (ML) plays a significant role in this perspective. Cancer diagnostic and prognosis microarray dataset is available with the biopsy dataset. Because of its importance in making diagnoses and classifying cancer diseases, the microarray data represents a massive amount. It may be challenging to do an analysis on a large number of datasets, though. As a result, feature selection is crucial, and machine learning provides classification techniques. These algorithms choose the relevant features that help build a more precise categorization model. Accurately classifying diseases is facilitated as a result, which aids in disease prevention. This work aims to synthesize existing knowledge on cancer diagnosis using machine learning techniques into a compact report. Current research work aims to propose an ensemble-based machine learning model En-PaFlower using Particle Swarm Optimization (PSO) as the feature selection algorithm, Flower Pollination algorithm (FPA) as the optimization algorithm with the majority voting algorithm. Finally, the performance of the proposed algorithm is evaluated over three different types of cancer disease datasets with accuracy, precision, recall, specificity, and F-1 Score etc as the evaluation parameters. The empirical analysis shows that the proposed methodology shows highest accuracy as 95.65%.
Article Details
References
M. Khalsan et al., "A Survey of Machine Learning Approaches Applied to Gene Expression Analysis for Cancer Prediction," in IEEE Access, vol. 10, pp. 27522-27534, 2022, doi: 10.1109/ACCESS.2022.3146312.
R. L. Siegel, K. D. Miller, N. S. Wagle, and A. Jemal, “Cancer statistics, 2023.” CA: A Cancer Journal for Clinicians, vol. 73, no. 1, pp. 17-48, 2023, doi: 10.3322/caac.21763.
W. Astuti and Adiwijaya, “Support vector machine and principal component analysis for microarray data classification.” Journal of Physics: Conference Series, vol. 971, p. 012003, 2018, doi: 10.1088/1742-6596/971/1/012003.
I. M. Zubair and B. Kim, "A Group Feature Ranking and Selection Method Based on Dimension Reduction Technique in High-Dimensional Data," in IEEE Access, vol. 10, pp. 125136-125147, 2022, doi: 10.1109/ACCESS.2022.3225685.
S. K. Senapati, M. Shrivastava and S. Mahapatra, "PCSVM: A Hybrid Approach using Particle Swarm Optimization and Cuckoo Search for Effective Cancer Diagnosis," 2023 2nd Edition of IEEE Delhi Section Flagship Conference (DELCON), Rajpura, India, 2023, pp. 1-5, doi: 10.1109/DELCON57910.2023.10127354.
K. R. Kavitha, A. V. Ram, S. Anandu, S. Karthik, S. Kailas and N. M. Arjun, "PCA-based gene selection for cancer classification," 2018 IEEE International Conference on Computational Intelligence and Computing Research (ICCIC), Madurai, India, 2018, pp. 1-4, doi: 10.1109/ICCIC.2018.8782337.
A. Pati, M. Parhi, B. K. Pattanayak, B. Sahu, and S. Khasim, “CanDiag: Fog Empowered Transfer Deep Learning Based Approach for Cancer Diagnosis,” Designs, vol. 7, no. 3, p. 57, Apr. 2023, doi: 10.3390/designs7030057.
A. Pati et al., “Breast Cancer Diagnosis Based on IoT and Deep Transfer Learning Enabled by Fog Computing,” Diagnostics, vol. 13, no. 13, p. 2191, Jun. 2023, doi: 10.3390/diagnostics13132191.
A. Pati et al., “Diagnose Diabetic Mellitus Illness Based on IoT Smart Architecture,” Wireless Communications and Mobile Computing, vol. 2022, p. e7268571, Aug. 2022, doi: https://doi.org/10.1155/2022/7268571.
Gawande, G. S. ., Kanwade, A. B. ., Deshmukh, A. B. ., Bhandari, S. ., Bendre, V. ., & Dakre, A. G. . (2023). IoT-based Weather Information Using WeMos. International Journal of Intelligent Systems and Applications in Engineering, 11(3s), 85–92. Retrieved from https://ijisae.org/index.php/IJISAE/article/view/2534
A. Pati, M. Parhi, and B. K. Pattanayak, “A review on prediction of diabetes using machine learning and data mining classification techniques,” International Journal of Biomedical Engineering and Technology, vol. 41, no. 1, p. 83, 2023, doi: https://doi.org/10.1504/ijbet.2023.128514.
A. Pati, A. Panigrahi, D. S. K. Nayak, G. Sahoo, and D. Singh, “Predicting Pediatric Appendicitis using Ensemble Learning Techniques,” Procedia Computer Science, vol. 218, pp. 1166–1175, 2023, doi: https://doi.org/10.1016/j.procs.2023.01.095.
W. Jia, M. Sun, J. Lian, and S. Hou, “Feature dimensionality reduction: a review,” Complex & Intelligent Systems, Jan. 2022, doi: https://doi.org/10.1007/s40747-021-00637-x.
J. Zhu, “Classification of gene microarrays by penalized logistic regression,” Biostatistics, vol. 5, no. 3, pp. 427–443, Jul. 2004, doi: https://doi.org/10.1093/biostatistics/kxg046
X. Zhou, K.-Y. Liu, and S. T. C. Wong, “Cancer classification and prediction using logistic regression with Bayesian gene selection,” Journal of Biomedical Informatics, vol. 37, no. 4, pp. 249–259, Aug. 2004, doi: https://doi.org/10.1016/j.jbi.2004.07.009.
H. Zhao, S. Qi and Q. Dong, "Predicting prostate cancer progression with penalized logistic regression model based on co-expressed genes," 2012 5th International Conference on BioMedical Engineering and Informatics, Chongqing, China, 2012, pp. 976-980, doi: 10.1109/BMEI.2012.6512948.
F. Morais-Rodrigues et al., “Analysis of the microarray gene expression for breast cancer progression after the application modified logistic regression,” Gene, vol. 726, p. 144168, Feb. 2020, doi: https://doi.org/10.1016/j.gene.2019.144168.
[1]L. Fan, K.-L. Poh, and P. Zhou, “A sequential feature extraction approach for naïve bayes classification of microarray data,” Expert Systems with Applications, vol. 36, no. 6, pp. 9919–9923, Aug. 2009, doi: https://doi.org/10.1016/j.eswa.2009.01.075.
M. -Y. Wu, D. -Q. Dai, Y. Shi, H. Yan and X. -F. Zhang, "Biomarker Identification and Cancer Classification Based on Microarray Data Using Laplace Naive Bayes Model with Mean Shrinkage," in IEEE/ACM Transactions on Computational Biology and Bioinformatics, vol. 9, no. 6, pp. 1649-1662, Nov.-Dec. 2012, doi: 10.1109/TCBB.2012.105.
S. Nagi and D. Kr. Bhattacharyya, “Classification of microarray cancer data using ensemble approach,” Network Modeling Analysis in Health Informatics and Bioinformatics, vol. 2, no. 3, pp. 159–173, Jun. 2013, doi: https://doi.org/10.1007/s13721-013-0034-x.
M. A. Mahfouz, A. Shoukry, and M. A. Ismail, “EKNN: Ensemble classifier incorporating connectivity and density into kNN with application to cancer diagnosis,” Artificial Intelligence in Medicine, vol. 111, p. 101985, Jan. 2021, doi: https://doi.org/10.1016/j.artmed.2020.101985.
U. Maulik and D. Chakraborty, "Fuzzy Preference Based Feature Selection and Semisupervised SVM for Cancer Classification," in IEEE Transactions on NanoBioscience, vol. 13, no. 2, pp. 152-160, June 2014, doi: 10.1109/TNB.2014.2312132.
Y. Huo, L. Xin, C. Kang, M. Wang, Q. Ma, and B. Yu, “SGL-SVM: A novel method for tumor classification via support vector machine with sparse group Lasso,” Journal of Theoretical Biology, vol. 486, p. 110098, Feb. 2020, doi: https://doi.org/10.1016/j.jtbi.2019.110098.
N. Almugren and H. M. Alshamlan, "New Bio-Marker Gene Discovery Algorithms for Cancer Gene Expression Profile," in IEEE Access, vol. 7, pp. 136907-136913, 2019, doi: 10.1109/ACCESS.2019.2942413.
J. Cervantes, F. Garcia-Lamont, L. Rodríguez-Mazahua, and A. Lopez, “A comprehensive survey on support vector machine classification: Applications, challenges and trends,” Neurocomputing, vol. 408, pp. 189–215, Sep. 2020, doi: https://doi.org/10.1016/j.neucom.2019.10.118.
Maria Gonzalez, Machine Learning for Anomaly Detection in Network Security , Machine Learning Applications Conference Proceedings, Vol 1 2021.
B. Dai, R. -C. Chen, S. -Z. Zhu and W. -W. Zhang, "Using Random Forest Algorithm for Breast Cancer Diagnosis," 2018 International Symposium on Computer, Consumer and Control (IS3C), Taichung, Taiwan, 2018, pp. 449-452, doi: 10.1109/IS3C.2018.00119.
H. -J. Chiu, T. -H. S. Li and P. -H. Kuo, "Breast Cancer–Detection System Using PCA, Multilayer Perceptron, Transfer Learning, and Support Vector Machine," in IEEE Access, vol. 8, pp. 204309-204324, 2020, doi: 10.1109/ACCESS.2020.3036912.
Z. Guo, L. Xu, and N. Ali Asgharzadeholiaee, “A Homogeneous Ensemble Classifier for Breast Cancer Detection Using Parameters Tuning of MLP Neural Network,” Applied Artificial Intelligence, pp. 1–21, Jan. 2022, doi: https://doi.org/10.1080/08839514.2022.2031820.
A. Panigrahi, S. Bhutia, B. Sahu, M.G.Galety, and S.N. Mohanty, “BPSO-PSO-SVM: An Integrated Approach for Cancer Diagnosis,” pp. 571–579, Jan. 2022, doi: https://doi.org/10.1007/978-981-19-2177-3_53.
B. Sahu, A. Panigrahi, S. Pani, S. Swagatika, D. Singh and S. Kumar, "A Crow Particle Swarm Optimization Algorithm with Deep Neural Network (CPSO-DNN) for High Dimensional Data Analysis," 2020 International Conference on Communication and Signal Processing (ICCSP), Chennai, India, 2020, pp. 0357-0362, doi: 10.1109/ICCSP48568.2020.9182181.
P. E. Mergos and X.-S. Yang, “Flower pollination algorithm with pollinator attraction,” Evolutionary Intelligence, Jan. 2022, doi: https://doi.org/10.1007/s12065-022-00700-7.
Ólafur, S., Nieminen, J., Bakker, J., Mayer, M., & Schmid, P. Enhancing Engineering Project Management through Machine Learning Techniques. Kuwait Journal of Machine Learning, 1(1). Retrieved from http://kuwaitjournals.com/index.php/kjml/article/view/112
S. Lalljith, I. Fleming, U. Pillay, K. Naicker, Z. J. Naidoo and A. K. Saha, "Applications of Flower Pollination Algorithm in Electrical Power Systems: A Review," in IEEE Access, vol. 10, pp. 8924-8947, 2022, doi: 10.1109/ACCESS.2021.3138518.
O. A. Alomari, A. T. Khader, M. A. Al-Betar and Z. A. Alkareem Alyasseri, "A Hybrid Filter-Wrapper Gene Selection Method for Cancer Classification," 2018 2nd International Conference on BioSignal Analysis, Processing and Systems (ICBAPS), Kuching, Malaysia, 2018, pp. 113-118, doi: 10.1109/ICBAPS.2018.8527392.
B. Sahu and A. Panigrahi, “Efficient Role of Machine Learning Classifiers in the Prediction and Detection of Breast Cancer,” SSRN Electronic Journal, 2020, doi: https://doi.org/10.2139/ssrn.3545096.
B. Sahu, A. Panigrahi, S. K. Rout and A. Pati, "Hybrid Multiple Filter Embedded Political Optimizer for Feature Selection," 2022 International Conference on Intelligent Controller and Computing for Smart Power (ICICCSP), Hyderabad, India, 2022, pp. 1-6, doi: 10.1109/ICICCSP53532.2022.9862419.